 Zach has received a 2024 Evolutionary, Ecological, and Conservation Genomics (EECG) Research Award ($6,000) from the American Genetics Association for his project “Unveiling the mysteries of fog lichens: Phylogenomic insights into the phenotypic diversity of a niche-specific radiation. Congratulations!
Zach has received a 2024 Evolutionary, Ecological, and Conservation Genomics (EECG) Research Award ($6,000) from the American Genetics Association for his project “Unveiling the mysteries of fog lichens: Phylogenomic insights into the phenotypic diversity of a niche-specific radiation. Congratulations!
Author: Bernard Goffinet
Congratulations to Crystal awarded a SURF 2024
 Crystal (Zhu) was awarded a Summer Undergraduate Research Fellowship from the UConn Office of Undergraduate Research for her project “More than meets
Crystal (Zhu) was awarded a Summer Undergraduate Research Fellowship from the UConn Office of Undergraduate Research for her project “More than meets  the eye: Unveiling the diversity of lichen-forming fungal species in Chile using DNA barcoding”. The project focuses on Lobarioideae (Peltigerales) and primarily the genus Pseudocyphellaria. The award consists of a stipend and funds to cover research expenses. Congratulations.
the eye: Unveiling the diversity of lichen-forming fungal species in Chile using DNA barcoding”. The project focuses on Lobarioideae (Peltigerales) and primarily the genus Pseudocyphellaria. The award consists of a stipend and funds to cover research expenses. Congratulations.
The SURF program is jointly sponsored by the Office of Undergraduate Research, the Provost’s Office, the Office of the Vice President for Research, the Honors Program, donors to the university, and the Deans of the Schools and Colleges.
Crystal will be traveling to subantarctic Chile in May to see the various species in the Nothofagus forests on Navarino Island and near Punta Arenas.
New publication on mosses
 Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google Scholar
Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023. Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. pdf Google ScholarNew publication on bryophytes
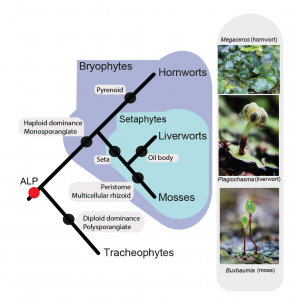 Bechteler J., G. Peñaloza-Bojacá, D. Bell, G. Burleigh, S. F. McDaniel, E. C. Davis, E. B. Sessa, A. Bippus, D. Christine Cargill, S. Chantanoarrapint, I. Draper, L. Endara, L. L. Forrest, R. Garilleti, S. W. Graham, S. Huttunen, J. Jauregui Lazo, F. Lara, J. Larraín, L. R. Lewis, D. G. Long, D. Quandt, K. Renzaglia, A. Schäfer-Verwimp, G. Ee Lee, A. S. Pinilla, M. von Konrat, C. E. Zartman, M. Regina Pereira, B. Goffinet & J. C. Villarreal. 2023. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. American Journal of Botany 110: e16249. pdf GoogleScholar
Bechteler J., G. Peñaloza-Bojacá, D. Bell, G. Burleigh, S. F. McDaniel, E. C. Davis, E. B. Sessa, A. Bippus, D. Christine Cargill, S. Chantanoarrapint, I. Draper, L. Endara, L. L. Forrest, R. Garilleti, S. W. Graham, S. Huttunen, J. Jauregui Lazo, F. Lara, J. Larraín, L. R. Lewis, D. G. Long, D. Quandt, K. Renzaglia, A. Schäfer-Verwimp, G. Ee Lee, A. S. Pinilla, M. von Konrat, C. E. Zartman, M. Regina Pereira, B. Goffinet & J. C. Villarreal. 2023. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. American Journal of Botany 110: e16249. pdf GoogleScholar
Abstract reads:
Premise: Bryophytes form a major component of terrestrial plant biomass, structuring ecological communities in all biomes. Our understanding of the evolutionary history of hornworts, liverworts and mosses has been significantly reshaped by inferences from molecular data, which have highlighted extensive homoplasy in various traits and repeated bursts of diversification. However, the timing of key events in the phylogeny, patterns and processes of diversification across bryophytes remain unclear.
Methods: Using the GoFlag probe set we sequenced 405 exons representing 228 nuclear genes for 531 species from 52 of the 54 orders of bryophytes. We inferred the species phylogeny from gene tree analyses using concatenated and coalescence approaches, assessed gene conflict, and estimated the timing of divergences based on 29 fossil calibrations.
Results: The phylogeny resolves many relationships across the bryophytes, enabling us to resurrect five liverwort orders and recognize three more, and propose ten new orders of mosses. Most orders originated in the Jurassic and diversified in the Cretaceous or later. The phylogenomic data also highlight topological conflict in parts of the tree, suggesting complex processes of diversification that cannot be adequately captured in a single gene-tree topology.
Conclusions: We sampled hundreds of loci across a broad phylogenetic spectrum spanning at least 450 Ma of evolution; these data resolved many of the critical nodes of the diversification of bryophytes. The data also highlight the need to explore the mechanisms underlying the phylogenetic ambiguity at specific nodes. The phylogenomic data provide an expandable framework toward reconstructing a comprehensive phylogeny of this important group of plants.
New publication on mosses
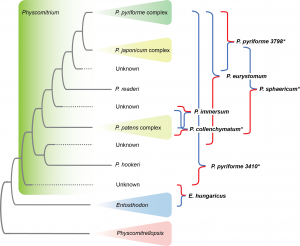 Patel N., R. Medina, L.D. Williams, O. Lemieux, B. Goffinet & M.G. Johnson. Frequent allopolyploidy with distant progenitors in the moss genera Physcomitrium and Entosthodon (Funariaceae) identified via subgenome phasing of targeted nuclear genes. Evolution 77: 2561–2575. pdf
Patel N., R. Medina, L.D. Williams, O. Lemieux, B. Goffinet & M.G. Johnson. Frequent allopolyploidy with distant progenitors in the moss genera Physcomitrium and Entosthodon (Funariaceae) identified via subgenome phasing of targeted nuclear genes. Evolution 77: 2561–2575. pdf
Abstract reads: Allopolyploids represent a new frontier in species discovery among embryophytes. Within mosses, allopolyploid discovery is challenged by low morphological complexity. The rapid expansion of sequencing approaches in addition to computational developments to identifying genome merger and whole-genome duplication using variation among nuclear loci representing homeologs has allowed for increased allopolyploid discovery among mosses. Here, we test a novel approach to phasing homeologs within loci and phasing loci across subgenomes, or subgenome assignment, called Homologizer, in the family Funariaceae. We confirm the intergeneric hybrid nature of Entosthodon hungaricus, and the allopolyploid origin of Physcomitrium eurystomum and of one population of P. collenchymatum. We also reveal that hybridization gave rise to P. immersum, as well as to yet unrecognized lineages sharing the phenotype of P. pyriforme, and P. sphaericum. Our findings demonstrate the utility of our approach when working with polyploid genomes, and its value in identifying progenitor species using target capture data.
New publication on lichens
Magain N., J. Miadlikowska, B. Goffinet, T. Goward, I. Juriado, A. Simon, C.J. Pardo De la Hoz, J. Mercado-Diaz, T. Barlow, B. Moncada, R. Lücking, A. Spielmann, L. Canez, L.S. Wang, P. Nelson, T. Wheeler, F. Lutzoni & E. Sérusiaux. 2023. High species richness in the lichen genus Peltigera (Ascomycota, Lecanoromycetes): 34 species in the dolichorhizoid and scabrosoid clades of sect. Polydactylon, including 23 new to science. Persoonia 51: 1–88. pdf GoogleScholar
Abstract reads: Applying molecular methods to fungi establishing lichenized associations with green algae or cyanobacteria has repeatedly revealed the existence of numerous phylogenetic taxa overlooked by classical taxonomic approaches. Here, we report taxonomical conclusions based on multiple species delimitation and validation analyses performed on an eight-locus dataset that includes world-wide representatives of the dolichorhizoid and scabrosoid clades in section Polydactylon of the genus Peltigera. Following the recommendations resulting from a consensus species delimitation approach and additional species validation analysis (BPP) performed in this study, we present a total of 25 species in the dolichorhizoid clade and nine in the scabrosoid clade, including respectively 18 and six species that are new to science and formally described. Additionally, one combination and three varieties (including two new to science) are proposed in the dolichorhizoid clade. The following 24 new species are described: P. appalachiensis, P. asiatica, P. borealis, P. borinquensis, P. chabanenkoae, P. clathrata, P. elixii, P. esslingeri, P. flabellae, P. gallowayi, P. hawaiiensis, P. holtanhartwigii, P. itatiaiae, P. hokkaidoensis, P. kukwae, P. massonii, P. mikado, P. nigriventris, P. orientalis, P. rangiferina, P. sipmanii, P. stanleyensis, P. vitikainenii and P. willdenowii; the following new varieties are introduced: P. kukwae var. phyllidiata and P. truculenta var. austroscabrosa; and the following new combination is introduced: P. hymenina var. dissecta. Each species from the dolichorhizoid and scabrosoid clades is morphologically and chemically described, illustrated, and characterised with ITS sequences. Identification keys are provided for the main biogeographic regions where species from the two clades occur. Morphological and chemical characters that are commonly used for species identification in the genus Peltigera cannot be applied to unambiguously recognise most molecularly circumscribed species, due to high variation of thalli formed by individuals within a fungal species, including the presence of distinct morphs in some cases, or low interspecific variation in others. The four commonly recognised morphospecies: P. dolichorhiza, P. neopolydactyla, P. pulverulenta and P. scabrosa in the dolichorhizoid and scabrosoid clades represent species complexes spread across multiple and often phylogenetically distantly related lineages. Geographic origin of specimens is often helpful for species recognition; however, ITS sequences are frequently required for a reliable identification.
New publication on mosses
 Hu R., X. Li, Y. Hu, R. Zhang, Q. Lv, M. Zhang, X. Sheng, F. Zhao, Z. Chen, Y. Ding, H. Yuan, X. Wu, S. Xing, X. Yan, F. Bao, P. Wan, L. Xiao, X. Wang, W. Xiao, E.L. Decker, N. van Gessel, H. Renault, G. Wiedemann, N.A. Horst, F.B. Haas, P.K.I. Wilhelmsson, K. Ullrich, E. Neumann, B. Lv, C. Liang, H. Du, H. Lu, Q. Gao, Z. Cheng, H. You, P. Xin, J. Chu, C.-H. Huang, Y. Liu, S. Dong, L. Zhang, F. Chen, L. Deng, F. Duan, W. Zhao, K. Li, Z. Li, X. Li, H. Cui, Y. Zhang, C. Ma, R. Zhu, Y. Jia, M. Wang, M. Hasebe, J. Fu, B. Goffinet, H. Ma, S.A. Rensing, R. Reski, Y. He. 2023. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell 186: 3558–3576. pdf
Hu R., X. Li, Y. Hu, R. Zhang, Q. Lv, M. Zhang, X. Sheng, F. Zhao, Z. Chen, Y. Ding, H. Yuan, X. Wu, S. Xing, X. Yan, F. Bao, P. Wan, L. Xiao, X. Wang, W. Xiao, E.L. Decker, N. van Gessel, H. Renault, G. Wiedemann, N.A. Horst, F.B. Haas, P.K.I. Wilhelmsson, K. Ullrich, E. Neumann, B. Lv, C. Liang, H. Du, H. Lu, Q. Gao, Z. Cheng, H. You, P. Xin, J. Chu, C.-H. Huang, Y. Liu, S. Dong, L. Zhang, F. Chen, L. Deng, F. Duan, W. Zhao, K. Li, Z. Li, X. Li, H. Cui, Y. Zhang, C. Ma, R. Zhu, Y. Jia, M. Wang, M. Hasebe, J. Fu, B. Goffinet, H. Ma, S.A. Rensing, R. Reski, Y. He. 2023. Adaptive evolution of the enigmatic Takakia now facing climate change in Tibet. Cell 186: 3558–3576. pdf
The most extreme environments are the most vulnerable to transformation under a rapidly changing climate. These ecosystems harbor some of the most specialized species, which will likely suffer the highest extinction rates. We document the steepest temperature increase (2010–2021) on record at altitudes of above 4,000 m, triggering a decline of the relictual and highly adapted moss Takakia lepidozioides. Its de-novo-sequenced genome with 27,467 protein-coding genes includes distinct adaptations to abiotic stresses and comprises the largest number of fast-evolving genes under positive selection. The uplift of the study site in the last 65 million years has resulted in life-threatening UV-B radiation and drastically reduced temperatures, and we detected several of the molecular adaptations of Takakia to these environmental changes. Surprisingly, specific morphological features likely occurred earlier than 165 mya in much warmer environments. Following nearly 400 million years of evolution and resilience, this species is now facing extinction.
New publication on bryophytes
Abstract reads: Physcomitrium pygmaeum is an ephemeral moss described in 1871 from a single collection from Utah, currently considered conspecific with Physcomitrium pyriforme. The interpretation of the taxon has been problematic due to its rarity in the field, the elusiveness of the type material, and an extremely scattered and inconsistent collection record. Here we present a comprehensive description and assessment of the taxon following the identification of the original material and lectotype designation, the examination of all existing herbarium specimens to the best of our knowledge, the collection of fresh material in Nevada, and the molecular barcoding of the latter using four plastid and two nuclear loci. Available information, albeit scant, suggests that this member of the North American bryoflora should be considered critically endangered following IUCN criteria.
New publication on bryophytes
New publication on bryophytes
Abstract reads: Background and Aims — With some 7,300 extant species, liverworts (Marchantiophyta) represent one of the major land plant lineages. The backbone relationships, such as the phylogenetic position of Ptilidiales, and the occurrence and timing of whole genome duplications, are still contentious. Methods – Based on analyses of the newly generated transcriptome data for 38 liverworts and complemented with those publicly available, we reconstructed the evolutionary history of liverworts and inferred gene duplication events along the 55-taxon liverwort species tree. Key Results – Our phylogenomic study provided an ordinal-level liverwort nuclear phylogeny and identified extensive gene tree conflicts and cyto-nuclear incongruences. Gene duplication analyses based on integrated phylogenomics and Ks distributions indicated no evidence of whole genome duplication events along the backbone phylogeny of liverworts. Conclusions – With a broadened sampling of liverwort transcriptomes, we re-evaluated the backbone phylogeny of liverworts, and provided evidence for ancient hybridizations followed by incomplete lineage sorting that shaped the deep evolutionary history of liverworts. The lack of whole genome duplication along the deep evolution of liverworts indicates that liverworts might represent one of the few major embryophyte lineages whose evolution was not driven by whole genome duplications.